M.I. Qadir1 and S.A. Malik2 1College of Pharmacy, GC University, Faisalabad, Pakistan.
2Department of Biochemistry, Faculty of Biological Sciences, Quaid-i-Azam University, Islamabad, Pakistan.
Address correspondence to:
M. Imran Qadir College of Pharmacy GC UniversityFaisalabad, Pakistan E-mail: mrimranqadir@hotmail.com
================================================== ======
Abstract
HIV fusion inhibitors may be classified into three groups. Peptides binding to HR1 include T1249, C30, and T20 (enfuvirtide). Peptides binding to HR2 include 5-helix. XTT formazan, NB-2, and NB-64 are nonpeptide fusion inhibitors. Genotypic testing for drug resistance is used more commonly than phenotypic testing because of its lower cost, wider availability, and shorter turnaround time. The aim of the study was to predict the efficacy of fusion inhibitors for AIDS patients in our population. A total of 100 specimens were collected. The viral RNA was isolated and nucleotides of the required regions were sequenced using the BigDye terminator method. It is concluded from this study that viruses in our population may show resistance to C30 and T20 whereas the other fusion inhibitors may be effectively used for our population.
Introduction
Drugs affecting the formation of the gp41 6-helix of HIV and ultimately inhibiting the fusion of the HIV membrane with human cells are classified as HIV fusion inhibitors.1 The HIV life cycle is started by fusion of the viral and human CD4 cellular membranes with the subsequent delivery of the viral genome into the host cell. Viral envelope glycoproteins play a critical role in the entry process.2,3 gp160 is translated from the env gene, which gives rise to gp120 and gp41.4 During the entry process, gp120 binds to its receptor CD4, resulting in conformational changes leading to dissociation of gp120 from gp41. Now the fusion peptide sequence of gp41 is inserted into the CD4 cell membrane. The next step is the folding of gp41 to form a 6-helix bundle, which is composed of three pairs of heptad repeat 1 and heptad repeat 2 (HR1 and HR2).5,6 This folding results in the closeness and finally the fusion of the HIV envelop and human CD4 cell membrane.7 Formation of the 6-helix bundle may be restricted by fusion inhibitors. HIV fusion inhibitors may be classified into three groups. Peptides binding to HR1 include T1249, C30, and T20 (enfuvirtide). Peptides binding to HR2 include 5-helix. XTT formazan, NB-2, and NB-64 are nonpeptide fusion inhibitors.1
Drug resistance may be evaluated phenotypically or genotypically. The genotypic estimation of drug resistance has more advantages than phenotypic estimation. Genotypic testing of drug resistance cost s less than phenotypic testing. Furthermore, genotypic testing has wider availability and a shorter turnaround time. Early evidence of drug resistance may also be predicted by genotypic assays. Genotypic testing identifies mutations even if the mutation occurs at a level too low to influence susceptibility in phenotypic testing, and detects transitional mutations that do not cause drug resistance by themselves but indicate the presence of selective drug pressure.8–11
The aim of the present study was to investigate what genetic variation is present in the HR1 and HR2 regions of the env gene of HIV strains. Does the genetic variability affect the amino acid sequences of the resulting protein? Mutations in the HR regions interfere with the fusion property of the virus membrane with human cells. Amino acid substitutions in the HR regions of gp41 may affect the efficacy of fusion inhibitors.12–14 The major aim of this study was to predict the efficacy of HIV fusion inhibitors for our population.
Materials and Methods
One hundred HIV blood samples were collected from the AIDS patients. These patients were enrolled by the National AIDS Control Program of Pakistan. Viral RNA was extracted from plasma samples using the QIAmp viral RNA kit (Qiagen, Valencia, CA). Reverse transcription polymerase chain reaction (RT-PCR) was then performed using primers designed to amplify a 589-bp region within gp41 that includes both HR1 and HR2 domains. The outside primers were L5′gp41 (TTCAGACCTGGAGGAGGAGATA) and L3′Env (GGTGGTAGCTGAAGAGGCACAGG). First-round amplification was performed using 50 pmol each of L5′gp41 and L3′Env. PCR conditions were 2min at 94°C, followed by 35 cycles of 30s at 94°C, 30s at 50°C, and 1min at 72°C, with an extension of 5min at 72°C. Second-round amplification was performed using the same protocol as above, but with primers L5′HR1 (AGAAGAGTGGTGCAGAGAGAAAA) and L3′HR2 (GGTGAGTATCCCTGCCTAACTCT). The purified nested PCR products from the gp41-amplified region were used for automated sequencing with a BigDye terminator cycle-sequencing ready reaction kit (PE Applied Biosystems, Foster City, CA) according to the manufacturer's protocol.
The chromatograms obtained from the affected individuals were compared with one another by developing a phylogenetic tree. The trees were developed by using PHYLIP (Phylogeny Inference Package) version 3.5c by Joseph Felsenstein, Department of Genetics, University of Washington, Seattle, available at
http://hiv.lanl.gov.
Results
Nucleotide sequence of HR1
A common nucleotide sequence of the HR1 region of the env gene of HIV was 7382ACGGTACAGGCCAGACGATTATTGTCTGGTATAGTGCAGCAGCAGA ACAATTTGCTGAGGGCTATTGAGGCGCAACAGCATCTGTTGCAACTCACA GTCTGGGGCATCAAGCAGCTCCAG7501. This nucleotide sequence results in the following amino acid sequence: 538TVRARRLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQ577. Several mutations were present in the nucleotide sequence of the HR1 region of the env gene of HIV. These mutations resulted in amino acid shifts at three positions: R543Q, R557K, and Q567R. A summary of the amino acid sequence of the HR1 region is given in Fig. 1.
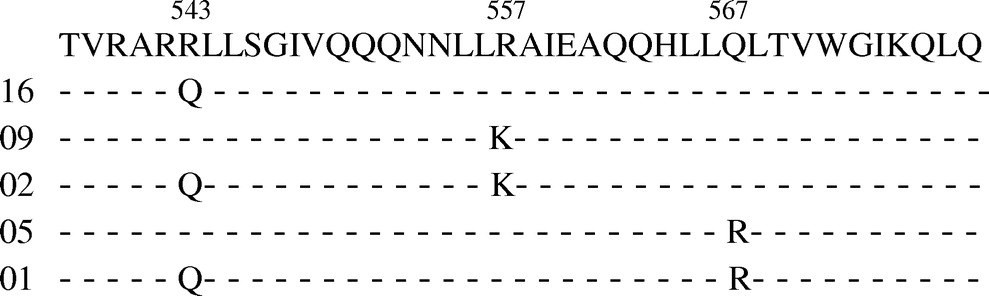 FIG. 1.
FIG. 1. A summary of the amino acid sequence of the HR1 region of the env gene of HIV specimens. The first numerical row shows the position of amino acids that have variations in our specimens. The second row indicates the sequence of amino acids denoted by one letter symbols. The left column indicates the number of specimens that shows variations. Each variation is indicated by a single letter symbol and the rest of the sequence is represented by dashes (–).
Information about Downloading
Nucleotide sequence of HR2
A common nucleotide sequence of the HR2 region of the env gene of HIV was 7652TGGATGGAGTGGGACAGAGAAATTAACAATTACACAACCTTACTAC ACTCCTTAATTGAAGAAGCGCAAAACCAGCAAGAAAAGAATGAACAAGAC TTATTGGAATTAGATAAATGGGCAAGTTTGTGGAATTGGTTT7789. This nucleotide sequence results in the following amino acid sequence: 628WMEWDREINNYTTLLHSLIEEAQNQQEKNEQDLLELDKWASLWNWF6 73. Several mutations were also present in the nucleotide sequence of the HR2 region of the env gene of HIV. These mutations resulted in amino acid shifts at three positions: M629R, N636D, T640S, T640N, S644T, A649S, D659E, S668N, and N671S. Only a single silent mutation was found: C7690T resulting in T640T.
A summary of the amino acid sequence of the HR2 region is given in Fig. 2. The amino acid sequences of HR1 and HR2 of gp41 of wild-type HIV along with the change in amino acids from the wild type obtained in our study are given in Fig. 3.
 FIG. 2.
FIG. 2. A summary of the amino acid sequence of the HR2 region of the env gene of HIV specimens. The first numerical row shows the position of amino acids that have variations in our specimens. The second row indicates the sequence of amino acids denoted by one letter symbols. The left column indicates the number of specimens that shows variations. Each variation is indicated by a single letter symbol and the rest of the sequence is represented by dashes (–).
Information about Downloading
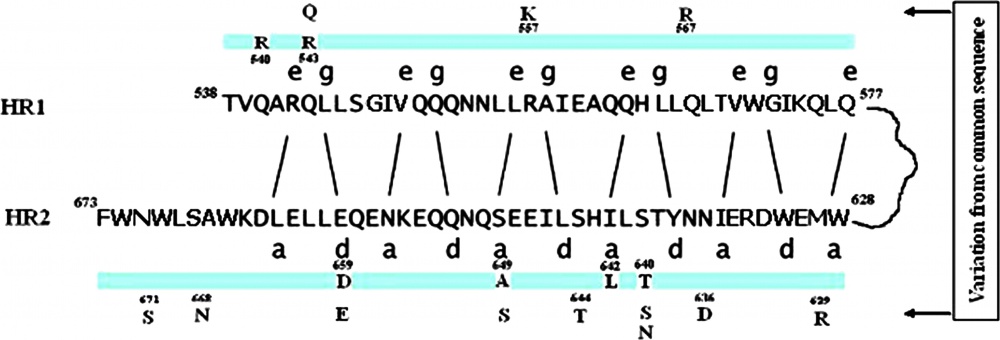 FIG. 3.
FIG. 3. Amino acids sequences of HR1 and HR2 of gp41 of wild-type HIV. Hydrophobic interactions between position e of HR1 and position a of HR2 and between position g of HR1 and position d of HR2 are present. The change in amino acids in HR regions from the wild-type HIV obtained in this study is given in the ribbon with the position of the amino acid on its cap. Variation from the “common sequence obtained in this study” is given outside the ribbon.
Information about Downloading
Phylogenetic analysis
The phylogenetic tree for HR1 is given in Fig. 4. With reference to HR1, viruses in our specimens may be divided into six groups. The phylogenetic tree of HR2 is given in Fig. 5. With reference to HR2, viruses in our specimens have greater diversity and may be divided into 17 groups.
 FIG. 4.
FIG. 4. Phylogenetic tree of the HR1 region of the env gene of HIV made with the neighbor-joining method and an F84 distance model with equal site rates, with 100 sequences and 120 characters including insertions and gaps stripped. Made on
www.hiv.lanl.gov.
Information about Downloading
 FIG. 5.
FIG. 5. Phylogenetic tree of the HR2 region of the env gene of HIV made with the neighbor-joining method and an F84 distance model with equal site rates, with 100 sequences and 138 characters including insertions and gaps stripped. Made on
www.hiv.lanl.gov.
Information about Downloading
Discussion
Before discussing the interaction between the amino acids of the HR regions and their respective drugs, we should be aware of the interactions that stabilize the 6-helix bundle. A disulfide bond may be present between two cysteine residues. The carboxyl group containing a negative charge in the side chain in aspartate (D) and glutamate (E) can interact with the positive charged group in the side chain such as the amino group of lysine (K). Amino acid side chains containing oxygen- or nitrogen-bound hydrogen can form hydrogen bonds with electron-rich atoms at the side chain.15 In this study, hydrophobicity is responsible for the amino acid interactions, except for those between Q551 and Q652, which are based on hydrogen bonding. Young et al.16 and Jones and Thornton17 also proposed a hydrophobic interaction between position e of HR1 and position a of HR2 and between position g of HR1 and position d of HR2. The hydrophobic interaction between position a and position d of the adjacent HR1 stabilizes the central trimeric core.
Peptides binding to HR1 includes T1249, C30, and T20 (enfuvirtide). Thus, mutations at position e or g in HR1 may involve resistance to these drugs.18 542R, 544L, 549V, 551Q, 556L, 558A, 563Q, 565L570V, 572G, and 577Q of HR1 are the binding amino acids for T1249. 544L, 549V, 551Q, 556L, 558A, 563Q, 565L, 570V, 572G, and 577Q of HR1 are the binding amino acids for C30 whereas 542R, 544L, 549V, 551Q, 556L, 558A, 563Q, and 565L of HR1 are the binding amino acids for T20.1 HR1 of gp41 is a highly conserved region.19 Although the V549A mutational change was reported to be the most resistance-associated mutation in HR1,20 no single case of this mutation has been seen in our studies. We did, however, find some other mutations in the leucine zipper of the HR1 with reference to the wild-type sequence. These include 540R (100%), 543R (81%), 557R (89%), and 567Q (94%). These mutations were not present at either the e or the g position, which may be involved in the interaction with T1249, C30, or T20.
Amino acids of HR2 showed greater variation than amino acids of HR1. The S640T, I642L, S649A, and E659D mutations, from the wild-type virus, were seen in our study. The first mutation is at the f position, and therefore it does not interact with amino acids of HR1. The middle two mutations at the a position and the last at the d position will interact with amino acids of HR1. The I642L and E659D mutations do not affect the hydrophobicity, but the S649A mutation leads to a shift from a hydrophilic amino acid to a hydrophobic amino acid. Thus the stability of the 6-helix bundle will be increased as the hydrophobic amino acid is present at the HR1 interacting position (556L). An S649A substitution was present in less than 5% of the observed cases by different workers.21,22 But in our case 88% of patients have the viruses with 649A. The fusion inhibitors (C30 and T20) bearing hydrophilic S at a position comparable to 649 in HR2 will not be able to compete with the hydrophobic attraction between A649 and L556 of the viruses. Therefore 6-helix will be stabilized and the viruses will show resistance to the drugs. However, a fusion inhibitor (T1249) that bears hydrophobic A at a position comparable to 649 in HR2 will compete with the hydrophobic attraction between A649 and L556.
The binding site for 5-helix is composed of amino acids present at position a or d of HR2, which include 628W, 631W, 635I, 638Y, 642I, 645L, 649S, 652Q, 656N, 659E, and 663L in the wild-type sequence. These amino acids also interact with position e or g of the HR1. In our study, limited variability was found among HR2 epitopes important for 5-helix interaction and binding. No significant published data related to in vivo resistance are available. However, analyses of the critical epitopes involved in the 5-helix interaction have been undertaken. In the present study, we examined the variability of these critical epitopes for 5-helix action and have found remarkable conservation at most of these critical positions, despite a high variability in the overall HR2 domain.
Of the 11 critical residues identified, nine had identical residues across all specimens tested, and the remaining two residues at position 649 and 659 contained variations from our common sequence. The variations observed from our common sequence were at position 649, an amino acid change from A→S, and at position 659, an amino acid change from D→E. This varied sequence was actually present in the wild-type virus.23 The common critical sequence for 5-helix interaction observed by Hanna et al.22 was the same as in the wild type. However, there were some samples with variations S649A and E659D and these variations constitute our common sequence. Moreover, the I635V and L645I mutations related to the 5-helix interaction were also observed in Hanna's study.
Both amino acid changes lead to a decrease in hydrophobicity that may affect the binding capability of the 5-helix. In addition to S649A and E659D, one more amino acid change was also present in our common sequence as compared to the wild-type sequence and this includes I642L. A shift of I642L and S649A leads to a clear increase in hydrophobicity, whereas an E659D shift causes a slight decrease in hydrophobicity, resulting in an overall increase in hydrophobicity at the binding positions of the 5-helix. Therefore it may be concluded that the 5-helix will have greater binding ability with the HR2 region of HIV in our population as compared to the wild-type virus and will be a very strong antiviral drug candidate for HIV in our population.
XTT formazan is a nonpeptide fusion inhibitor that binds to the central hydrophobic core in the trimeric coiled-coil region of gp41. The amino acids at the a or d position of HR1 are involved in the construction of the central hydrophobic core. No amino acid changes at position a or d of HR1 was seen in this study. Therefore, for our population, XTT formazan may act in the same way it acts for other populations.
K574 is highly conserved24 and no substitution was observed at K574 in our study. Therefore, it may be concluded that drugs binding to K574 such as NB-2 and NB-64 will also be effective for our population.
Conclusions
It is concluded from the present study that mutations in the nucleotide sequences of HR regions also affect the amino acid sequences of the resulting protein. Only one silent mutation was found within the hundred specimens. A fusion inhibitor (T1249) that bears hydrophobic A at a position comparable to 649 in HR2 will be able to compete with the hydrophobic attraction between A649 and L556 and thus may be effectively used for our population. But the fusion inhibitors (C30 and T20) bearing hydrophilic S at a position comparable to 649 in HR2 will not be able to compete with the hydrophobic attraction between A649 and L556 of the viruses. Therefore 6-helix will be stabilized and the viruses will show resistance to the drugs. This resistance in our population may be avoided if the hydrophilic S of these fusion inhibitors, at a position comparable to 649 in HR2, is replaced by hydrophobic A (Qadir-C30 and Qadirvirtide).
Mutations in the HR2 region of HIV in our population result in an overall increase in hydrophobicity at the binding positions of the 5-helix. Therefore it may be concluded that 5-helix will have more binding ability with the HR2 region of HIV in our population as compared to the wild-type virus and will be a very strong antiviral drug candidate for HIV in our population. Nonpeptide compounds such as XTT formazan, NB-2, and NB-64 may be effectively used for our population as no mutations were found related to their binding sites.
Sequence Data
The sequences discussed in this study were deposited into GenBank under accession numbers HM436821–HM437020 for env gp41.
Acknowledgments
We are thankful to Prof. Mazhar Hayat and M. Asif, Department of English, GC University, Faisalabad, Pakistan for their kind help in improving this article.
References
1. Qadir MI and Malik SA: HIV fusion inhibitors. Rev Med Virol 2010;20:23–33.
2. Lewis P, Hensel M, and Emerman M: Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J 1992;11:3053–3058.
3. Lassen KG, Lobritz MA, Bailey JR, Johnston S, and Nguyen S: Elite suppressor-derived HIV-1 envelope glycoproteins exhibit reduced entry efficiency and kinetics. PLoS Pathog 2009;5(4):e1000377.
4. McCune JM, Rabin LB, Feinberg MB, Lieberman M, Kosek JC, Reyes GR, and Weissman IL: Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 1988;53:55–67.
5. Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, and Wiley DC: Atomic structure of the ectodomain from HIV-1 gp41. Nature 1997;387:426–430.
6. Qadir MI, Malik SA, Nisa TU, Tabassum N, Ali S, and Nisar L: Characterization of HR region of gp41 of HIV. Int J Agric Biol 2010;12:456–458.
7. Chan DC, Fass D, Berger JM, and Kim PS: Core structure of gp41 from the HIV envelope glycoprotein. Cell 1997;89:263–273.
8. Baxter JD, Mayers DL, Wentworth DN, Neaton JD, Hoover ML, Winters MA, Mannheimer SB, Thompson MA, Abrams DI, Brizz BJ, Ioannidis JP, and Merigan TC: A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS 2000;14:F83–F93.
9. Cingolani A, Antinori A, Rizzo MG, Murri R, Ammassari A, Baldini F, Di Giambenedetto S, Cauda R, and De Luca A: Usefulness of monitoring HIV drug resistance and adherence in individuals failing highly active antiretroviral therapy: A randomized study (ARGENTA). AIDS 2002;16(3):369–379.
10. Meynard JL, Vray M, Morand-Joubert L, Race E, Descamps D, Peytavin G, Matheron S, Lamotte C, Guiramand S, Costagliola D, Brun-Vizinet F, Clavel F, and Girard P-M: Phenotypic or genotypic resistance testing for choosing antiretroviral therapy after treatment failure: A randomized trial. AIDS 2002;16:727–736.
11. Tural C, Ruiz L, Holtzer C, Schapiro J, Viciana P, Gonzalez J, Domingo P, Boucher C, Rey-Joly C, and Clotet B: Clinical utility of HIV-1 genotyping and expert advice: The Havana trial. AIDS 2002;16(2):209–218.
12. Ivanoff LA, Dubay JW, Morris JF, Roberts SJ, Gutshall L, Sternberg EJ, Hunter E, Matthews TJ, and Petteway SR Jr: V3 loop region of the HIV-1 gp120 envelope protein is essential for virus infectivity. Virology 1992;187(2):423–432.
13. Wolf E: HIV drug resistance. 2005.
http://www.hivmedicine.com/textbook/rt.html, retrieved on 11-09-09.
14. Wolf E: HIV resistance testing. In: HIV Medicine 2006 (Hoffmann C, Rockstroh JK, and Kamps BS, Eds.). Flying Publisher, Paris, 2006, pp. 331–356.
15. Champe PC and Harvey RA: Structure of proteins. In: Lippincott's Illustrated Review: Biochemistry, 2nd ed. J.B. Lippincott Company, Philadelphia, 1994, pp. 13–24.
16. Young L, Jernigan RL, and Covell DG: A role for surface hydrophobicity in protein-protein recognition. Protein Sci 1994;3:717–729.
17. Jones S and Thornton JM: Principles of protein-protein interactions. Proc Natl Acad Sci USA 1996;93:13–20.
18. Kazmierski WM, Kenakin TP, and Gudmundsson KS: Peptide, peptidomimetic and small-molecule drug discovery targeting HIV-1 host-cell attachment and entry through gp120, gp41, CCR5 and CXCR4. Chem Biol Drug Des 2006;67:13–26.
19. Melby T, Heilek G, Cammack N, and Greenberg ML: Resistance to enfuvirtide and other HIV entry inhibitors. In: Antimicrobial Drug Resistance (Mayers DL, Ed.). Humana Press, LLC, Totawa, NJ, 2009, pp. 493–506.
20. Sista PR, Melby T, Davison D, Jin L, Mosier S, Mink M, Nelson EL, DeMasi R, Cammack N, Salgo MP, Matthews TJ, and Greenberg ML: Characterization of determinants of genotypic and phenotypic resistance to enfuvirtide in baseline and on-treatment HIV-1 isolates. AIDS 2004;18:1787–1794.
21. Dorn J, Masciotra S, Yang C, Downing R, Biryahwaho B, Mastro TD, Nkengasong J, Pieniazek D, Rayfield MA, Hu DJ, and Lal RB: Analysis of genetic variability within the immunodominant epitopes of envelope gp41 from human immunodeficiency virus type 1 (HIV-1) group M and its impact on HIV-1 antibody detection. J Clin Microbiol 2000;38:773–780.
22. Hanna SL, Yang C, Owen SM, and Lai RB: Resistance mutation in HIV entry inhibitors. AIDS 2002;16:1603–1608.
23. Root MJ, Kay MS, and Kim PS: Protein design of an HIV-1 entry inhibitor. Science 2001;291:884–888.
24. Markosyan RB, Leung M, and Cohen FS: The six-helix bundle of HIV Env controls pore formation and enlargement and is initiated at residues proximal to the hairpin turn. J Virol 2009;83(19):10048–10057.






 Similar Threads
Similar Threads 

 .BZU.
.BZU. Don't cry because it's over, smile because it happened
Don't cry because it's over, smile because it happened 






 Linear Mode
Linear Mode

